Understanding Bayesian Updates with a Simple Gaussian Model
Bayesian inference is a framework for updating beliefs in light of new evidence. In this post we go through the formulas involved in estimating the mean of a gaussian dataset, along with code to do said process.
The Gaussian Distribution
A Gaussian (or normal) distribution is described by two parameters: the mean and standard deviation . Its probability density function (PDF) is given by:
In this example we generate synthetic data from a Gaussian distribution with a true mean and variance .
Using this data as our true data we then want to estimate by iteratively update our belief about using Bayesian inference.
We write this in python as:
import numpy as np
import matplotlib.pyplot as plt
# Define a Gaussian function
def gaussian(x, mean, std):
return (1 / (np.sqrt(2 * np.pi) * std)) * np.exp(-0.5 * ((x - mean) / std) ** 2)
and we generate a synthetic dataset from this gaussian
# Generate data by sampling from a Gaussian
np.random.seed(42)
true_mean, true_std = 5, 2
n_samples = 1000
x_values = np.linspace(true_mean - 4 * true_std, true_mean + 4 * true_std, 1000)
distribution = gaussian(x_values, true_mean, true_std)
data = np.random.choice(x_values, size=n_samples, p=distribution / np.sum(distribution))
The variable data is now a list of observations from that distribution.
Bayesian Framework
Bayes' theorem is a fundamental theorem in probability theory that describes how to update the probability of a hypothesis based on new evidence. It is expressed as:
Where:
- is the posterior probability, the probability of the hypothesis given the data.
- is the likelihood, the probability of the data given the hypothesis.
- is the prior probability, the initial probability of the hypothesis.
- is the marginal likelihood or evidence, the total probability of the data.
We can rewrite this as:
Why is Used
The use of (proportional to) arises because Bayes' theorem includes a normalization constant, the evidence :
This ensures the posterior is a valid probability distribution. However, is independent of , so when calculating the posterior shape, we can write:
This simplification:
- Avoids explicitly calculating , which can be computationally expensive.
- Focuses on the relative contributions of the prior and likelihood.
- Leaves normalization implicit, which is straightforward for Gaussian distributions.
Derivation of Posterior Updates
Prior and Likelihood
The prior and likelihood are both Gaussian distributions:
- Prior:
- Likelihood:
Posterior Distribution
Combining the prior and likelihood:
Expanding and grouping terms involving :
From this, we identify the posterior as a Gaussian distribution with updated parameters:
- Posterior Variance:
- Posterior Mean:
Iterative Updates with Multiple Observations
When observing multiple data points , the posterior is updated iteratively. After observations, the posterior parameters are:
- Variance:
- Mean:
This iterative process is implemented in the code, leading to convergence of the posterior mean and variance as more data is observed.
# Prior parameters
prior_mean, prior_var = 0, 10
# Likelihood parameters (assumed known)
likelihood_var = true_std**2
# Bayesian updates
posterior_mean, posterior_var = prior_mean, prior_var
means, vars = [], []
for i in range(n_samples):
posterior_mean = (posterior_mean / posterior_var + data[i] / likelihood_var) / ( 1 / posterior_var + 1 / likelihood_var )
posterior_var = 1 / (1 / posterior_var + 1 / likelihood_var)
means.append(posterior_mean)
vars.append(posterior_var)
posterior_std = np.sqrt(posterior_var)
print(f"True mean: {true_mean}")
print(f"Prior mean: {prior_mean} ± {np.sqrt(prior_var)}")
print(f"Posterior mean: {posterior_mean} ± {posterior_std}")
Results and Visualization
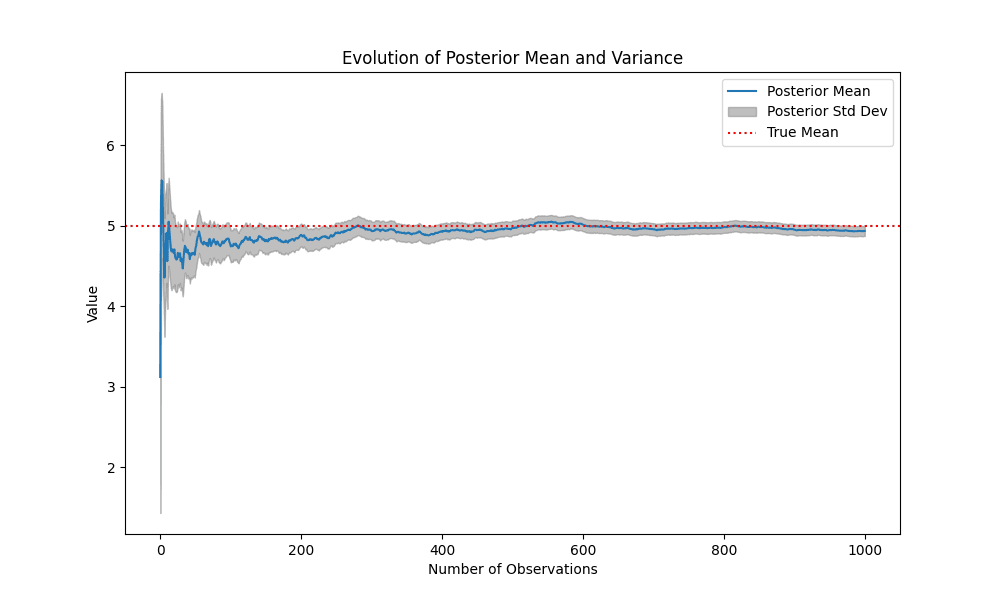
The Python implementation shows that as , the posterior parameters approach the true values:
Visualization:
The plot below illustrates the evolution of the posterior mean and variance over time:
- Posterior Mean: Converges to the true mean ().
- Posterior Variance: Shrinks, indicating increased confidence.
Full code:
import numpy as np
import matplotlib.pyplot as plt
# Define a Gaussian function
def gaussian(x, mean, std):
return (1 / (np.sqrt(2 * np.pi) * std)) * np.exp(-0.5 * ((x - mean) / std) ** 2)
# Generate data by sampling from a Gaussian
np.random.seed(42)
true_mean, true_std = 5, 2
n_samples = 1000
x_values = np.linspace(true_mean - 4 * true_std, true_mean + 4 * true_std, 1000)
distribution = gaussian(x_values, true_mean, true_std)
data = np.random.choice(x_values, size=n_samples, p=distribution / np.sum(distribution))
# Prior parameters
prior_mean, prior_var = 0, 10
# Likelihood parameters (assumed known)
likelihood_var = true_std**2
# Bayesian updates
posterior_mean, posterior_var = prior_mean, prior_var
means, vars = [], []
for i in range(n_samples):
posterior_mean = (posterior_mean / posterior_var + data[i] / likelihood_var) / ( 1 / posterior_var + 1 / likelihood_var )
posterior_var = 1 / (1 / posterior_var + 1 / likelihood_var)
means.append(posterior_mean)
vars.append(posterior_var)
posterior_std = np.sqrt(posterior_var)
# Results
print(f"True mean: {true_mean}")
print(f"Prior mean: {prior_mean} ± {np.sqrt(prior_var)}")
print(f"Posterior mean: {posterior_mean} ± {posterior_std}")
# Plot posterior evolution
plt.figure(figsize=(10, 6))
plt.plot(means, label="Posterior Mean")
plt.fill_between(
range(n_samples),
np.array(means) - np.sqrt(vars),
np.array(means) + np.sqrt(vars),
color="gray",
alpha=0.5,
label="Posterior Std Dev",
)
plt.axhline(true_mean, color="red", linestyle="dotted", label="True Mean")
plt.title("Evolution of Posterior Mean and Variance")
plt.xlabel("Number of Observations")
plt.ylabel("Value")
plt.legend()
plt.savefig("posterior_evolution_gaussian.png")
plt.show()